Approved
Standard Operating Protocol for Investigating Data Quality at Sites
Purpose
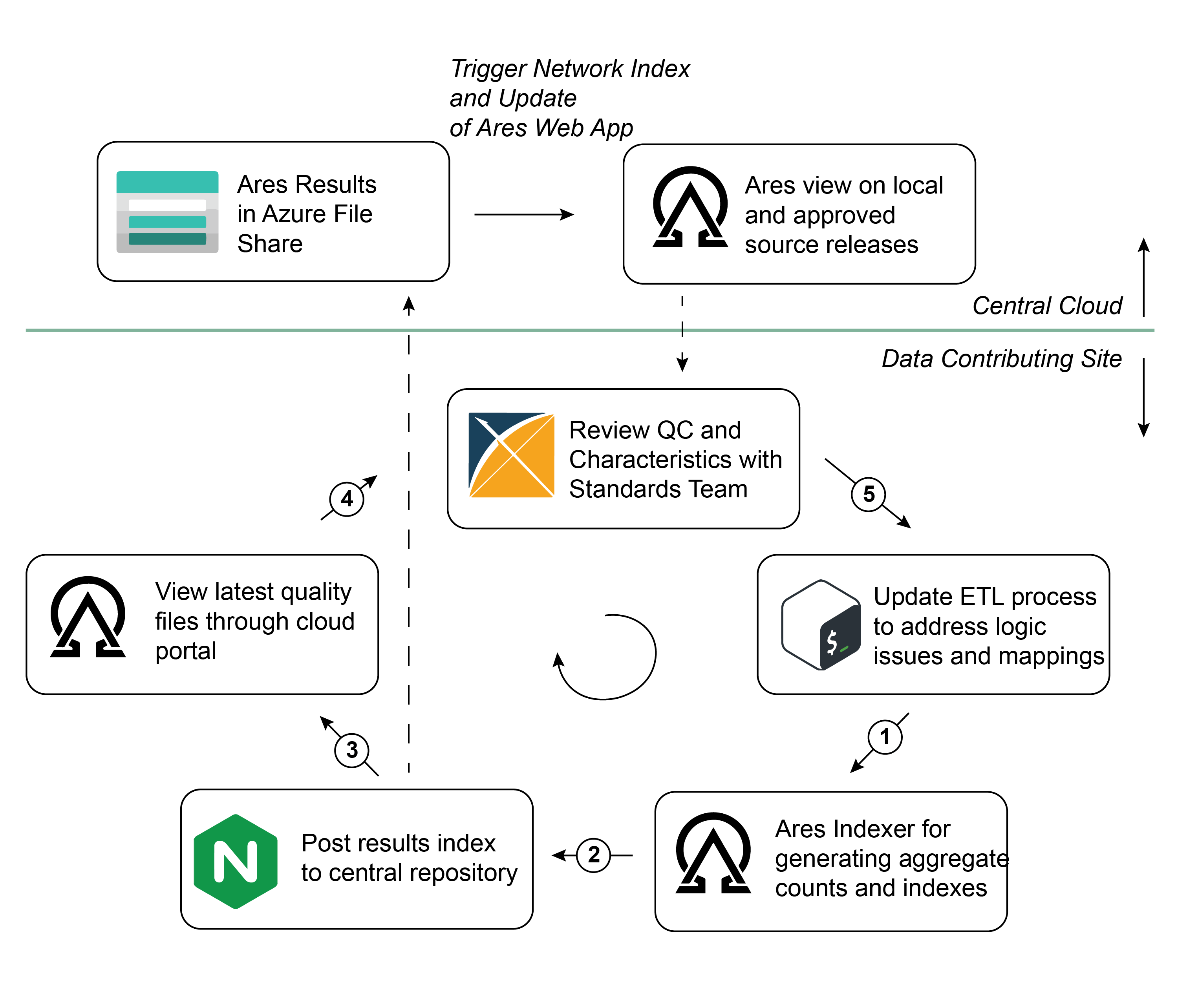
This SOP outlines a step-by-step approach for evaluating local data quality at sites, submitting quality overviews to the central location, and reviewing and updating an ETL implementation accordingly.
Note that the workflow below can be followed without submitting full data extracts to the central location. The only interaction with the central cloud that is necessary is the submission of the Ares Index files, which contain aggregate counts of concepts and data quality overviews.
We expect that this iterative workflow will be executed more frequently than the full data extracts will be submitted.
STEP 1: EXECUTE QC TOOLS LOCALLY
The first step in evaluating the quality and characteristics of your site's data locally is launching the QC tools against those data. There are three main tools we're focused on within CHoRUS, and that we plan to update in order to tailor to CHoRUS-specific needs as those needs become better defined.
These tools are:
- Achilles
- Achilles executes a variety of characterization-oriented queries against an OMOP dataset to extract patient counts per concept, prevalence over time, etc.
- Data Quality Dashboard - DQD
- DQD runs a variety of quality-oriented queries against an OMOP dataset to evaluate those data with regard to their completeness, conformance, and plausibility
- AresIndexer
- The AresIndexer augments output from the above tools to prepare it for visualization in the Ares web application
In order to execute these processes locally, you can either:
(A) configure a local R environment and run the relevant packages against your database
- Head to the OHDSI Hades documentation page and follow the R-setup instructions for your operating system
- Install the three packages and their associated dependencies above using the
remotespackage- e.g.
remotes::install_github("/OHDSI/DataQualityDashboard")
- e.g.
- Connect to your database and execute the packages in the following order
Achilles::achilles(...)DataQualityDashboard::executeDqChecks(...)Achilles::exportToAres(...)Achilles::performTemporalCharacterization(...)- Note that this function is not essential and may fail if you don't have concepts that meet its time constraints (36 months)
AresIndexer::augmentConceptFiles(...)
- Note that an example of the above process flow can be found in the chorus-container-apps repo
- Once you've executed these checks, you should have a data index directory with the following structure
(B) use the existing chorus-etl Docker image that has all components and dependencies installed and available
- pull the public
chorus-etlDocker imagedocker pull ghcr.io/chorus-ai/chorus-etl:main
- launch it either locally or in the cloud
- if on local machine:
- run the command
docker run -it --entrypoint /bin/bash --name chorus-etl -v /<some_local_dir>:/ares_output ghcr.io/chorus-ai/chorus-etl:main- This command launches a container based on the
chorus-etlimage and places you at a command line console for that container
- This command launches a container based on the
- install a command-line editor like nano
> apt update && apt install nano - update the
ares.Rfile in the container using the editor above to reference your database connection parameters- you will need to set the
aresDataRootparameter to/ares_output, which you mounted in the command above
- you will need to set the
- launch the file above
Rscript /opt/etl/src/etl/ares.R <args if desired>
- run the command
- if in data site's cloud:
- send an email to Jared to discuss details
- if on local machine:
- Once the checks have executed successfully, you should be able to see a dated data index in
<some_local_dir>that you configured in the Docker command above
Relevant Resources
OFFICE HOURS
- Achilles Output
- DQD Output
- DQD Overview
- Ares Overview
- Contributing Ares Results Pt 1
- Constructing Ares Index for Upload
OTHERS
STEP 2: ACCESS UPLOAD PORTAL AND SUBMIT RELEASE INDEX
One member from each Data Generating Site (DGS) will receive an external user access credential to the MGH Azure cloud environment. With this credential, you will be able to access the external cloud portal to upload results and view ares.
The DGS portal (scripted here and built into chorus-www-dgs) will have links to two locations:
-
An Ares web application instance with the following sources:
- Ingested Data Releases for Each DGS + MERGE
- Approved Data Releases for Each DGS + MERGE
- Remote Data Releases for Each DGS
-
An AresIndex upload portal, where you can upload a compressed (.zip) copy of your release index
- e.g.
20240203.zip
- e.g.
IF you would like to deploy the Ares web application locally for tracking data quality and characterization at your site, we have built a Docker image and provided documentation to get you started.
Relevant Resources
OFFICE HOURS
- Deploy Ares Web App Locally
- A Standards office hour session walking through this upload workflow is planned
STEP 3: REVIEW RECENTLY SUBMITTED RELEASE(S) IN ARES
As mentioned in STEP 2, a user from each DGS will have access to the external Ares portal. Here, that user will be able to review Ares results from their site and compare those results to other sites within the network.
Note that each DGS will appear three separate times in this Ares view:
- Releases that have been ingested in the cloud but not approved
- Releases that have been approved for integration into the CHoRUS analytics enclave
- Releases that describe the state of your CHoRUS data locally (created by you in STEP 1)
In your review, it's important to investigate and understand any issues in the data quality tab for your particular source release, and then link those issues with potential logic and/or mapping updates in a future ETL version.
Relevant Resources
OFFICE HOURS
STEP 4: MEET WITH STANDARDS TEAM TO EVALUATE RESULTS AND DEFINE NEXT STEPS
Marty Alvarez is the point of contact for scheduling one-on-one meetings with the Standards Team. Most DGS have already scheduled and participated in these meetings; we anticipate they will become very useful and targeted once DGS have uploaded data that can be discussed in detail.
These discussions will likely evaluate severity of quality checks, ability of DGS data to support downstream CHoRUS analytics, and priorities for updating the ETL and mappings appropriately.
It could be that a meeting is not necessary and the source release submission represents a data update with no logic changes rather than data stemming from new logic. If so, the DGS can skip STEP 4 and proceed to STEP 5.
STEP 5: IMPLEMENT PRIORITIZED UPDATES AND RE-RUN ETL
Following a meeting and review together with the Standards Team (should that be necessary), each DGS will need to work to implement any necessary changes to the ETL logic, and to update/validate any associated mappings.
This iterative update process will likely be multidisciplinary, requiring input from data engineers, clinicians, informaticists, or ontologists.
The process itself is essential in curating high-quality data extracts and, ultimately, supporting meaningful analytics downstream.
Following updates, the DGS can return to Step 1, re-executing the quality checks and proceeding
Related Office Hours
The following office hour sessions provide additional context and demonstrations related to this SOP:
-
[06-15-2023] Data Quality Dashboard (DQD)
- Video Recording | Transcript
- Introduction to Data Quality Dashboard tools and methodology
-
[07-06-2023] DQD Output Demo
- Video Recording | Transcript
- Demonstration of DQD output interpretation and analysis
-
[10-26-23] Review and Prioritization of DQD Results, and Discussion of DQD Issue Severity
- Video Recording | Transcript
- Detailed session on prioritizing and addressing DQD findings
-
[05-02-24] Using ARES to assess data readiness for the upcoming CHoRUS challenge
- Video Recording | Transcript
- Practical application of ARES for data quality assessment
-
[02-15-24] Constructing the Ares Index for Upload
- Video Recording | Transcript
- Technical guidance on ARES index construction and upload
-
[03-07-24] Deploying Ares locally and Mapping Data
- Video Recording | Transcript
- Local deployment and configuration of ARES tools